Optimizing Gene Selection: A Mini Review on Reference Gene Normalization for qRT-PCR in Solanaceae Plants
##plugins.themes.academic_pro.article.main##
Abstract
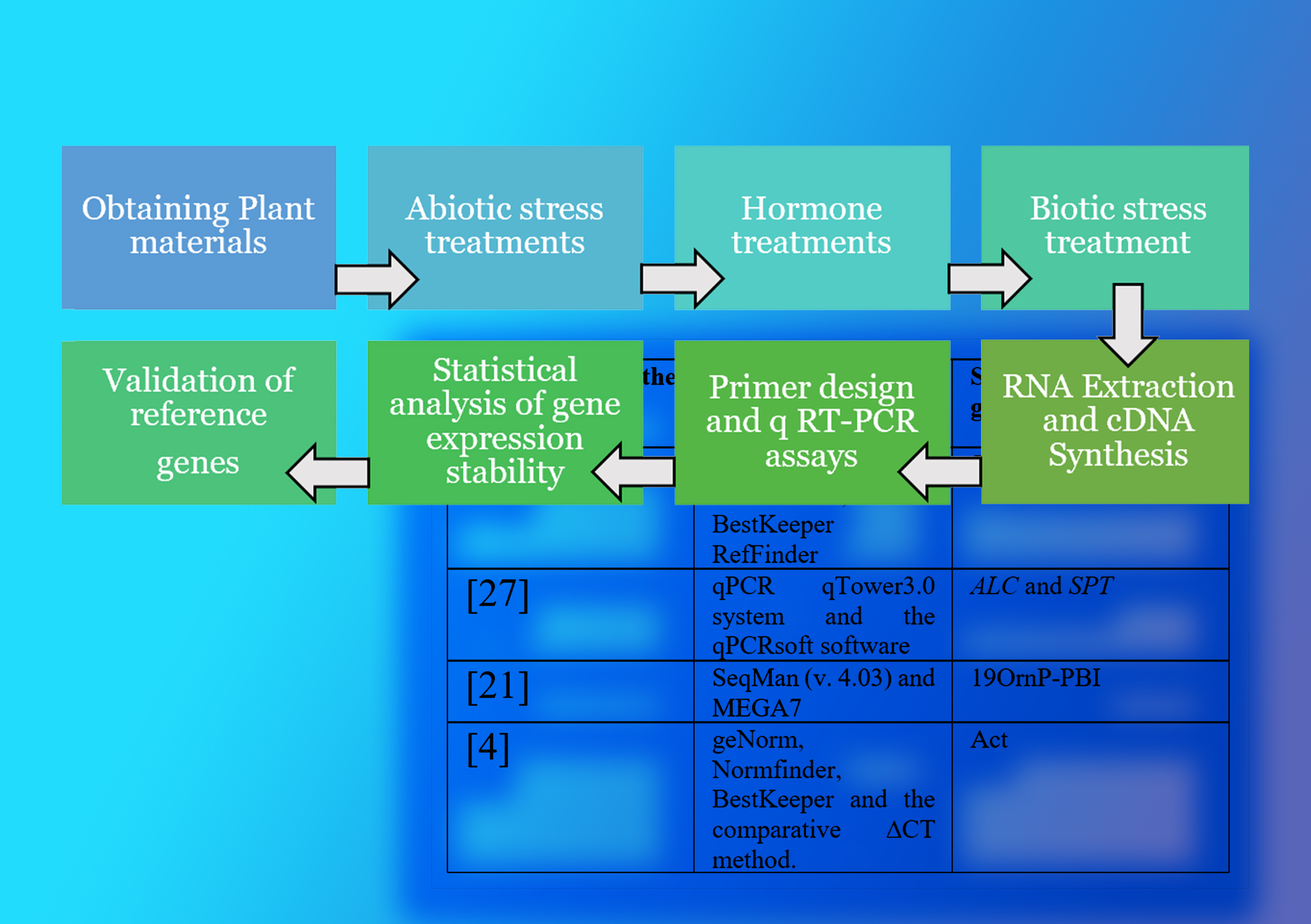
Gene expression analysis is fundamental for understanding biological processes, and quantitative real-time PCR (qRT-PCR) has become a widely used method for validating the expressions. Proper normalization across multiple samples and tissues is crucial for accurate and reliable results, requiring the selection of suitable reference genes for comparison with target genes. Finding internal reference genes in Solanaceae plants is crucial for precise transcript quantification using real-time PCR. Research on Solanaceae plants like Solanum rostratum, Solanum melongena, Solanum tuberosum, Solanum lycopersicum, Nicotiana tabacum, and Capsicum was conducted to identify reference genes for qRT-PCR normalization. Researchers can confidently assess gene expression variations in Solanaceae plants by establishing accurate reference genes, providing deeper insights into biological processes. This literature review provides a comprehensive overview of methodologies, challenges, and advancements in the selection of candidate reference genes for RT-PCR in Solanaceae plants. It has the potential to influence future research directions and methodologies, enhance scholarly discourse, and pursue excellence in molecular biology research.